Abstract
Refractoriness to induction chemotherapy and relapse after achievement of remission are the main barriers to cure in acute myeloid leukemia (AML). Currently, most AML patients receive standard induction chemotherapy because risk stratification depends on cytogenetic and molecular tests that typically take weeks to return results. We recently reported a rapid 17-gene leukemia stem cell score (LSC17, Ng et al., Nature 2016) that can accurately determine risk within 24 to 48 hours of diagnosis. Patients with high LSC17 scores have poor outcomes with current treatments and could be enrolled in clinical trials evaluating novel upfront treatment strategies.
A small subset of AML patients with normal cytogenetics, NPM1 mutation and no FLT3-ITD are classified as low molecular risk (LMR) and are considered to have better outcomes, but ~35% relapse within 2 years following conventional therapy. In our prior study, we derived an optimized, re-weighted sub-score for LMR patients in which only 3 of the 17 genes contribute to the calculated score (LSC3). A high LSC3 score identifies LMR patients who have worse outcomes following standard induction therapy, and who might benefit from novel frontline therapy. Currently however, the LSC3 score can only be employed for risk-adapted therapy decisions in the post-remission setting, as LMR patients are only identified following cytogenetic and molecular testing. Earlier identification of high-risk LMR patients will facilitate clinical trials of novel frontline therapy in this patient subset.
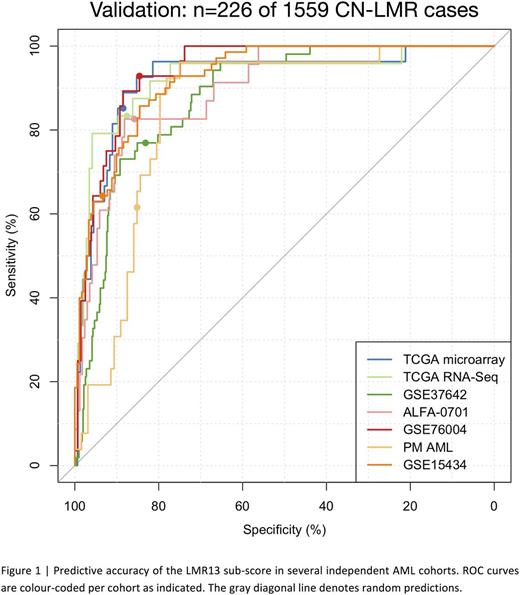
To determine whether a subset of the LSC17 signature genes could be used to identify LMR cases at diagnosis, we applied sparse binomial regression to relate the expression of the 17 signature genes to LMR vs. non-LMR cases in a large training cohort comprising AML patients of all subtypes (GSE6891, n=495 patients including 48 [9%] LMR cases). The resulting 13-gene sub-score (LMR13) can accurately identify LMR patients in 6 large independent validation cohorts spanning n=1559 cytogenetically and molecularly diverse AML patients using gene expression data from several different platforms (Figure 1; microarray datasets: GSE37642, n=542, area under the receiver operating characteristic curve (AUROC)=0.88; GSE15434, n=251, AUROC=0.92; TCGA AML, n=183, AUROC=0.92; ALFA-0701, n=192, AUROC=0.89; RNA-Seq dataset: TCGA AML, n=169, AUROC=0.92; NanoString datasets: GSE76004, n=237, AUROC=0.96; PM AML, n=154, AUROC=0.86). Importantly, the LMR13 score for each patient can be calculated from the same NanoString assay used to determine the patient's LSC17 and LSC3 risk scores. The ability to identify LMR cases rapidly at diagnosis enhances the clinical utility of the LSC17 assay, as it will allow clinicians to use the most appropriate risk score (LSC3 vs LSC17) for all newly-diagnosed AML patients, and enable evaluation of novel upfront treatment strategies for higher-risk LMR cases. Furthermore, integration of our functionally defined stem cell gene list with patient survival data using statistical learning approaches provides a basis for ongoing characterization of the molecular pathways associated with clinical outcomes, irrespective of mutational profile.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.